esm.doi.bio/esm3/esm3.biological_alignment.full1
==============================
We study how the base models can be aligned (40) to generate proteins that satisfy challenging prompts. To do this, for each model we construct a dataset of partial structure prompts, generate multiple protein sequences for each prompt, and then fold and score each of the sequences using ESM3 for consistency with the prompt (cRMSD) and foldability (pTM). High quality samples are paired with low quality samples for the same prompt to construct a preference dataset (Appendix A.4). ESM3 is then tuned to optimize a preference tuning loss, which incentivizes the model to put higher likelihood on the high quality samples compared to low quality samples (Appendix A.4) (41, 42).
We examine the ability of the model to generate highquality scaffolds using challenging tertiary motif scaffolding prompts. We prompt ESM3 with the amino acid identities and atomic coordinates of residues derived from a dataset of 46 ligand binding motifs in a set of temporally held out proteins (Appendix A.4.5). For each motif task, we create 1024 prompts by permuting the order of the residues, varying their position in the sequence, and varying the length of the sequence. A single protein is generated per prompt. We evaluate success using the percentage of tasks solved (backbone cRMSD $<1.5 \AA$, pTM $>0.8$ ) after 128 generations (Appendix A.4.5).
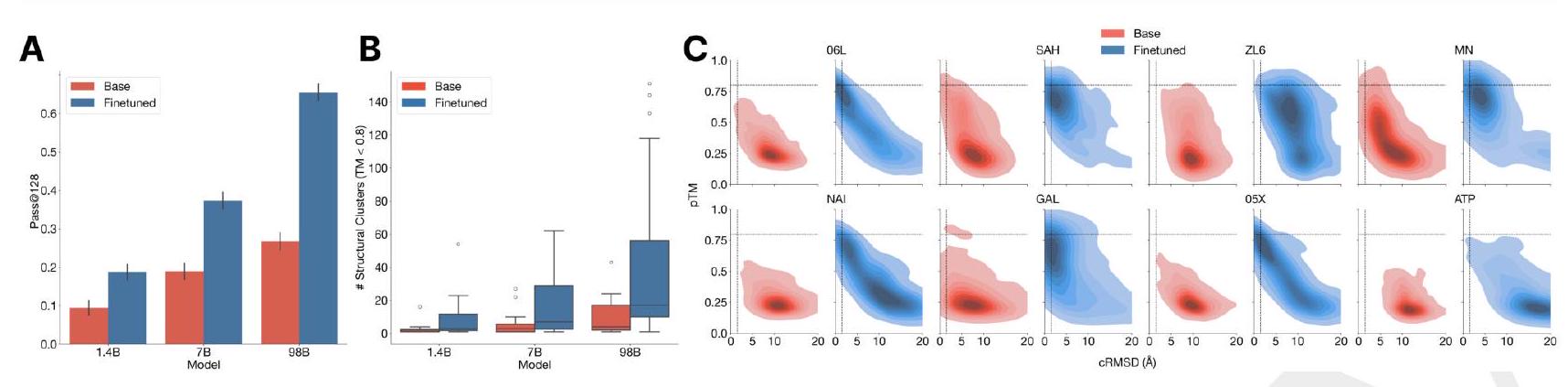
Preference tuned models solve double the atomic coordination tasks compared to base models (Fig. 3A). While the base models show differences in the fraction of tasks solved $(9.5 \%$ for 1.4B, $19.0 \%$ for 7B, 26.8\% for 98B; Fig. 3A), a much larger capability difference is revealed through align-
Preference tuned models solve double the atomic coordination tasks compared to base models (Fig. 3A). While the base models show differences in the fraction of tasks solved $(9.5 \%$ for 1.4B, $19.0 \%$ for 7B, 26.8\% for 98B; Fig. 3A), a much larger capability difference is revealed through alignment-based evaluation (Fig. 3B). The preference tuned models are much better at aligning to the correct structures, with a $2.5-3.5\times$ improvement in the fraction of tasks solved.
User:
ESM3
alignment
preference tuning loss
pTM
cRMSD
tertiary contact
tertiary motif
ligands
densities of prompted generations
User:
ment $(9.5 \%$ to $18.8 \%, 19.0 \%$ to $37.4 \%, 26.8 \%$ to $65.5 \%$ for the 1.4B, 7B and 98B models, respectively). Preferencetuned models not only solve a greater proportion of tasks, but also find a greater number of solutions per task, as evaluated by the number of distinct structural clusters ( $\mathrm{TM}>0.8$ ) with backbone cRMSD $<1.5$ Åand pTM $>0.8$ (Fig. 3B). A shift in the distribution of ESMFold pTM and backbone cRMSD for each ligand binding motif is observed (Fig. 3C; Fig. S17). At the 98B scale, the finetuned model produces more distinct successful clusters than the base model on 37 of the 46 tested ligands, while the remaining 9 ligands were not solved by either the base or aligned model, indicating that alignment almost universally improves the faithfulness to the prompt and the foldability of the generated proteins. Compared to a supervised finetuning baseline, which only maximizes the likelihood of the positive examples, preference tuning leads to larger improvements at all scales (Appendix A.4.6).
Obsidian Markdown Internal Link ESMFold TM pTM ligand binding motif ligand alignment supervised finetuning baseline positive examples Appendix A.4.6
User:
These results demonstrate that preference tuning extracts latent capability in the models. The capability of larger models to solve challenging tasks become far more apparent after alignment. Since alignment can be performed with arbitrary objectives, this is an indication of a general ability to respond to finetuning that greatly improves with scale.
These results demonstrate that preference tuning extracts latent capability in the models. The capability of larger models to solve challenging tasks become far more apparent after alignment. Since alignment can be performed with arbitrary objectives, this is an indication of a general ability to respond to finetuning that greatly improves with scale.
In this context, "preference tuning" and "alignment" are not obvious to a non-expert, so I will create internal links for them.
Preference tuning: preference tuning
Alignment: alignment

