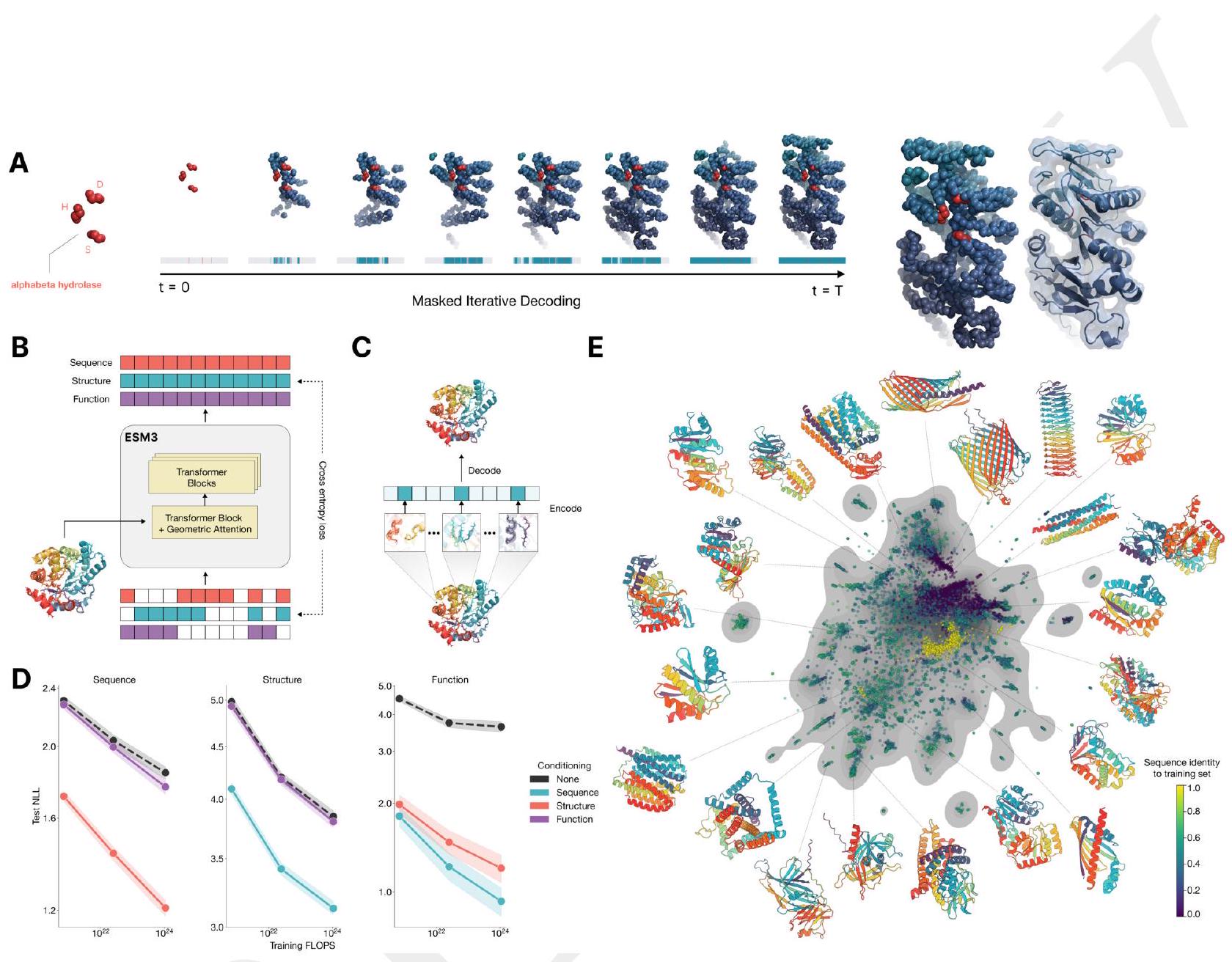
==============================
ESM3 reasons over the sequence, structure, and function of proteins. All three modalities are represented by tokens, and are input and output as separate tracks that are fused into a single latent space within the model. ESM3 is trained with a generative masked language modeling objective.
$$ \mathcal{L}=-\mathbb{E}{x, m}\left[\frac{1}{|m|} \sum{i \in m} \log p\left(x{i} \mid x{\backslash m}\right)\right]
A random mask $m$ is applied to the tokens $x$ describing the protein, and the model is supervised to predict the identity of the tokens that have been masked. During training, the mask is sampled from a noise schedule so that ESM3 sees many different combinations of masked sequence, structure, and function, and predicts completions of any combination of the modalities from any other. This differs from the classical masked language modeling (28) in that the supervision is applied across all possible masking rates rather than a single fixed masking rate. This supervision factorizes the probability distribution over all possible predictions of the next token given any combination of previous tokens, ensuring that tokens can be generated in any order from any starting point (29-31).
ESM3 masked language modeling noise schedule representation learning
Tokenization Protein structures Discrete auto-encoder Invariant geometric attention mechanism Local reference frames Global frame Computational primitives Attention Sequence of discrete tokens
ESM3 is a bidirectional transformer. While extensive research has gone into creating specialized architectures and training objectives for proteins, we find that tokenization paired with a standard masked language modeling objective and the basic transformer architecture is highly effective for both representation learning and generative modeling. Sequence, structure, and function tracks are input as tokens, which are embedded and fused, then processed through a transformer architecture.

Figure 1. ESM3 is a generative language model that reasons over the sequence, structure, and function of proteins. (A) Iterative sampling with ESM3. Sequence, structure, and function can all be used to prompt the model. At each timestep $\mathrm{t}$, a fraction of the masked positions are sampled until all positions are unmasked. (B) ESM3 architecture. Sequence, structure, and function are represented as tracks of discrete tokens at the input and output. The model is a series of transformer blocks, where all tracks are fused within a single latent space; geometric attention in the first block allows conditioning on atomic coordinates. ESM3 is supervised to predict masked tokens. (C) Structure tokenization. Local atomic structure around each amino acid is encoded into tokens. (D) Models are trained at three scales: 1.4B, 7B, and 98B parameters. Negative log likelihood on test set as a function of training FLOPs shows response to conditioning on each of the input tracks, improving with increasing FLOPs. (E) Unconditional generations from ESM3 98B (colored by sequence identity to the nearest sequence in the training set), embedded by ESM3, and projected by UMAP alongside randomly sampled sequences from UniProt (in gray). Generations are diverse, high quality, and cover the distribution of natural sequences.
ESM3 ESM3 architecture Structure tokenization Models are trained at three scales Unconditional generations from ESM3 98B
Scaling ESM3 from 1.4 billion to 98 billion parameters results in substantial improvements in the validation loss for all tracks, with the greatest improvements observed in sequence loss (Fig. 1D, Fig. S11). These gains in validation loss lead to better representation learning (Table S7 and Fig. S8). In single sequence structure prediction (Table S8) on CAMEO, ESM3 98B obtains 0.895 mean local distance difference test (LDDT) and surpasses ESMFold (0.865 LDDT). Unconditional generation produces high-quality proteins-with a mean predicted LDDT (pLDDT) 0.84 and predicted template modeling score (pTM) 0.52-that are diverse in both sequence (mean pairwise sequence identity 0.155 ) and structure (mean pairwise TM score 0.48 ), spanning the distribution of known proteins (Fig. 1E, Fig. S13).
ESM3 ESMFold CAMEO LDDT pLDDT pTM TM score