==============================
GFP chromophore cofactors substrates fluorescence jellyfish coral proteins genomes molecules cellular structures processes
This paragraph explains how proteins in the GFP family are responsible for the bright colors seen in jellyfish and coral. These proteins have a unique ability to create a fluorescent chromophore without needing any additional molecules or substances. This makes them very useful for scientists who want to label and track molecules, structures, or processes within cells or organisms. By inserting the GFP sequence into the genome of other organisms, researchers can create a visible marker that helps them study a wide range of biological phenomena.
GFP protein engineering natural world rational design machine learning high-throughput screening mutations amino acid coding sequence fluorescence high throughput experimentation
This paragraph discusses the GFP family, which has been the focus of protein engineering efforts for many years. Despite these efforts, most functional variants of GFP have been found in nature. However, scientists have been able to create new GFP sequences with improved properties through rational design and machine learning-assisted high-throughput screening. These new sequences typically have only a few mutations from the original sequence. In rare cases, scientists have been able to introduce up to 40-50 mutations while still retaining GFP fluorescence.
GFP autocatalytic process chromophore amino acids protein kinked central alpha helix eleven stranded beta barrel
GFP stands for Green Fluorescent Protein, which is a protein that emits green light when it is exposed to ultraviolet or blue light. The process of creating a new GFP involves a complex biochemical and physical process that results in the formation of a chromophore, which is responsible for the protein's fluorescence. This process is autocatalytic, meaning that it is self-sustaining and does not require any external factors to occur. The chromophore is formed from three key amino acids that are located in the core of the protein. The structure of GFP is unique, with a kinked central alpha helix surrounded by an eleven stranded beta barrel.

Obsidian is a note-taking app that allows users to create and organize notes using Markdown syntax. Markdown is a lightweight markup language that enables users to format text using simple symbols and characters. In Obsidian, users can create internal links between notes by enclosing the note title in double brackets, like this: Note Title. This creates a hyperlink that, when clicked, takes the user directly to the linked note.
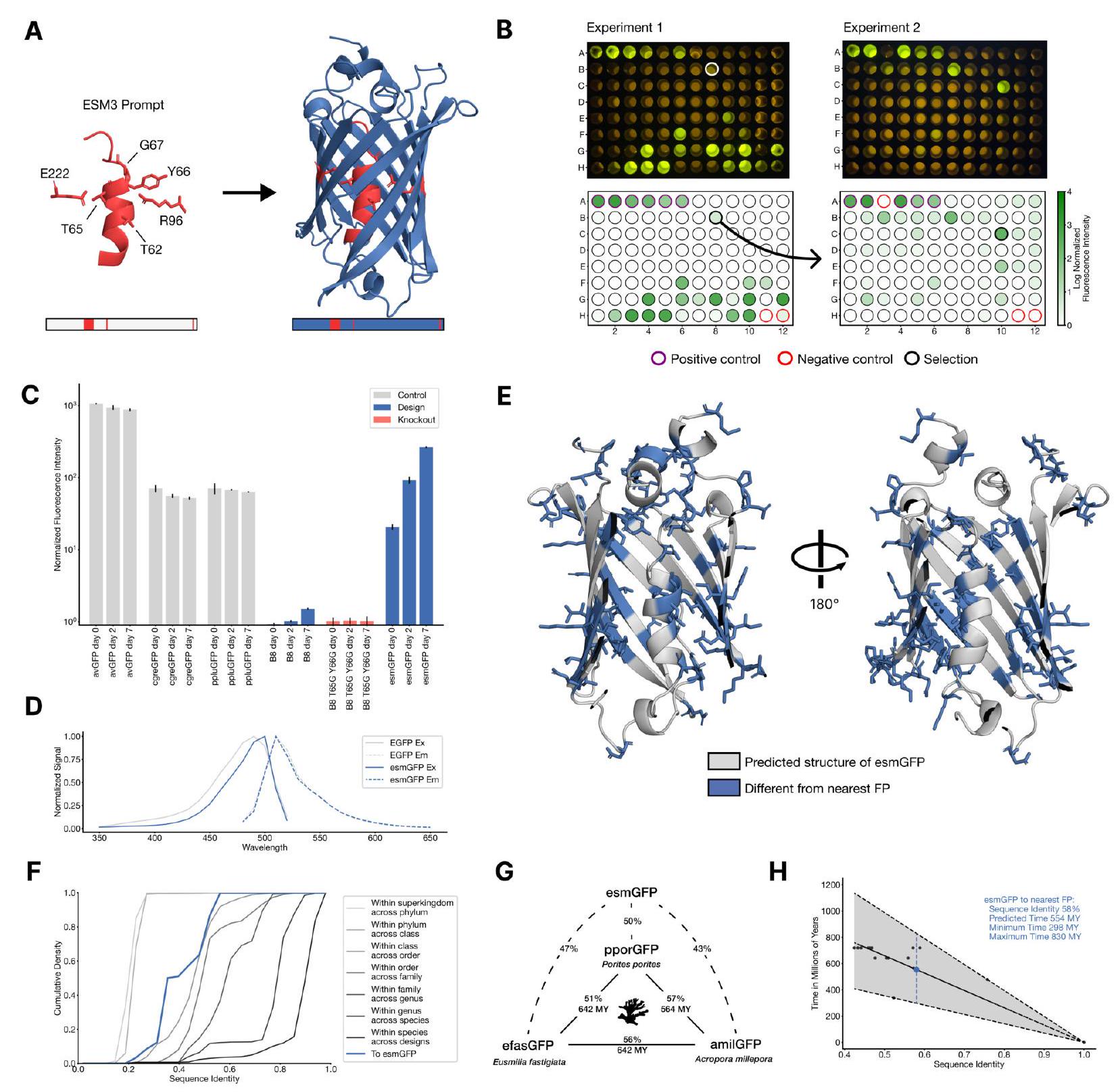
Figure 4. Generating a new fluorescent protein with a chain of thought. (A) We prompt ESM3 with the sequence and structure of residues required for forming and catalyzing the chromophore reaction, as well as the structure of part of the central alpha helix from a natural fluorescent protein (left). Through a chain of thought, ESM3 generates design candidates (right). (B) ESM3 found a bright GFP distant from other known GFPs in two experiments. We measured fluorescence in E. coli lysate. Top row, photograph of plates. Bottom row, plate reader fluorescence quantification. Positive controls of known GFPs are marked with purple circles, negative controls with no GFP sequence or no E. Coli are marked with red circles. In the first experiment (left) we expressed designs with a range of sequence identities. A notable design with low sequence identity to known fluorescent proteins appears in the well labeled B8 (highlighted in a black circle bottom, white circle top). We continue the chain of thought from the protein in B8 for the second experiment (right). A bright design appears in the well labeled C10 (black circle bottom, white circle top) which we designate esmGFP. (C) esmGFP exhibits fluorescence intensity similar to common GFPs. Normalized fluorescence is shown for a subset of proteins in experiment 2. (D) Excitation and emission spectra for esmGFP overlaid on the spectra of EGFP. (E) Two cutout views of the central alpha helix and the inside of the beta barrel of a predicted structure of esmGFP. The 96 mutations esmGFP has relative to its nearest neighbor, tagRFP, are shown in blue. (F) Cumulative density of sequence identity between fluorescent proteins across taxa. esmGFP has the level of similarity to all other FPs that is typically found when comparing sequences across orders, but within the same class. (G) Evolutionary distance by time in millions of years (MY) and sequence identities for three example anthozoa GFPs and esmGFP. (H) Estimator of evolutionary distance by time (MY) from GFP sequence identity. We estimate esmGFP is over 500 million years of natural evolution removed from the closest known protein.
In this paragraph, the authors discuss their process for generating a new fluorescent protein using ESM3, a computational tool. They explain that they prompted ESM3 with specific sequence and structure requirements for the chromophore reaction and part of the central alpha helix from a natural fluorescent protein. ESM3 then generated design candidates through a chain of thought. The authors tested these designs in E. coli lysate and found a bright GFP that was distant from other known GFPs. They designated this new protein as esmGFP and compared its fluorescence intensity and spectra to common GFPs. They also analyzed the sequence identity and evolutionary distance of esmGFP compared to other fluorescent proteins. Overall, the authors demonstrate the potential of ESM3 for generating new fluorescent proteins with precise configurations.
GFP ESM3 chromophore reaction Thr62 Thr65 Tyr66 Gly67 Arg96 Glu222 1QY3
In this paragraph, the researchers are trying to create new GFP sequences. They are using a program called ESM3 to generate a protein with 229 residues. They are focusing on specific critical residues that are important for forming and catalyzing the chromophore reaction. They are also using the structure of residues 58 through 71 from an experimental structure called 1QY3, which are known to be important for the energetic favorability of chromophore formation. They are providing sequence tokens, structure tokens, and atomic coordinates of the backbone as input for the program to generate the new GFP sequences.
GFP designs sequence optimization backbone structure tokens active site coordination iterative joint optimization candidate bucket similarity filter rank metrics
In this paragraph, the author is describing a process for generating designs of a protein called GFP. They start by creating a backbone structure using a chain-of-thought procedure, which involves generating structure tokens. These tokens are used to create a protein backbone that has good atomic coordination of the active site and is differentiated from the original backbone. The generated structure is then added to the original prompt to generate a sequence that is conditioned on the new prompt. The sequence and structure are then optimized iteratively, and any designs that lose atomic coordination of the active site are rejected. Finally, a computational pool of candidate GFP designs is drawn from the intermediate and final points in the iterative joint optimization stage. These designs are then bucketed by sequence similarity to known fluorescent proteins and filtered and ranked using a variety of metrics.
88 designs 96 well plate sequence similarity bucket synthesized expressed in E. coli measured for fluorescence activity excitation wavelength of $485 \mathrm{~nm}$ Fig. 4B left brightness positive controls naturally occurring GFPs sequence identity 1QY3 sequence tagRFP 50x less bright chromophore matured under a day new portion of sequence space protein engineering
In this paragraph, the researchers conducted an experiment with 88 different designs of proteins on a 96 well plate. They synthesized each protein and expressed it in E. coli, then measured its fluorescence activity at an excitation wavelength of $485 \mathrm{~nm}$. They found that some of the designs had similar brightness to positive controls, which are naturally occurring GFPs. They also identified a design in well B8 that had a lower sequence identity to the 1QY3 sequence and the nearest existing fluorescent protein, tagRFP. This design was less bright than natural GFPs and took longer to mature, but it showed potential for function in a new portion of sequence space that has not been found in nature or through protein engineering.
sequence design iterative joint optimization and ranking procedure 96 well plate plate reader assay brightness GFPs nature best design well C10 esmGFP
In this paragraph, the author is discussing the process of creating a protein with improved brightness. They start by using a sequence of design in well B8 and then use an iterative joint optimization and ranking procedure to generate a second 96 well plate of designs. They then use a plate reader assay to test the brightness of each design and find that several designs have a brightness similar to GFPs found in nature. The best design is located in well C10 of the second plate and is called esmGFP.
esmGFP B8 chromophore knockout of B8 avGFP cgreGFP ppluGFP Thr65 Tyr66 glycine
In this paragraph, the author is discussing the brightness of a protein called esmGFP compared to other similar proteins. They found that esmGFP takes longer to mature, but eventually becomes just as bright as the other proteins. They also tested whether certain amino acids were necessary for the protein to fluoresce, and found that when those amino acids were mutated, the protein lost its ability to fluoresce.
EsmGFP is a type of protein that has been analyzed in terms of its excitation and emission spectra. The excitation spectrum of EsmGFP has a peak at $496 \mathrm{~nm}$, which is slightly different from the peak of another protein called EGFP, which is at $489 \mathrm{~nm}$. However, both proteins emit at a peak of $512 \mathrm{~nm}$. The shape of the spectra shows that EsmGFP has a narrower full-width half-maximum (FWHM) for its excitation spectrum, but a highly comparable FWHM for its emission spectrum compared to EGFP. Overall, EsmGFP has spectral properties that are similar to other known GFPs.
esmGFP tagRFP eqFP578 BLAST MMseqs chromophore proximity interactions
In this paragraph, the author is discussing how they used different search methods to compare the sequence and structure of a protein called esmGFP to other known proteins. They found that the closest match to esmGFP was a protein called tagRFP, which was designed by scientists. The closest naturally occurring protein to esmGFP was eqFP578, which is a red fluorescent protein. The author also notes that there are many differences between the sequences of esmGFP and tagRFP, and that some of these differences occur in the interior of the protein, which is known to be very sensitive to mutations.
Examination of a sequence alignment of 648 natural and designed GFP-like fluorescent proteins revealed that esmGFP
GFP FP scleractinia actiniaria anthozoa hydrozoa avGFP
This paragraph discusses the similarity between different types of fluorescent proteins, specifically focusing on a new protein called esmGFP. The author notes that esmGFP is similar to other fluorescent proteins found in marine invertebrates, such as corals and sea anemones, but also shares some similarities with proteins found in jellyfish. The author uses a sequence alignment to show that esmGFP is more similar to proteins found in corals and anemones, with an average sequence identity of 51.4%, compared to proteins found in jellyfish, with an average sequence identity of 33.4%.
We can draw insight from evolutionary biology on the amount of time it would take for a protein with similar sequence identity to arise through natural evolution. In Fig. 4G we show esmGFP alongside three Anthozoan GFPs. We use a recent time-calibrated phylogenetic analysis of the Anthozoans (53) that estimated the millions of years ago (MYA) to last common ancestors to estimate evolutionary time between each pair of these species. Using a larger dataset of six Anthozoan GFPs and species for which we have accurate MYA to last common ancestors and GFP sequence identities, we construct a simple estimator that correlates sequence identity between FPs to MY of evolutionary time between the species (Fig. $4 \mathrm{H}$ ) to calibrate against natural evolution. Based on this analysis we estimate esmGFP represents an equivalent of over 500 million years of evolution from the closest protein that has been found in nature.
evolutionary biology natural evolution sequence identity Anthozoan GFPs time-calibrated phylogenetic analysis last common ancestors estimator sequence identity MY of evolutionary time natural evolution
In this paragraph, the authors are discussing how they used evolutionary biology to estimate the amount of time it would take for a protein with similar sequence identity to arise through natural evolution. They used a time-calibrated phylogenetic analysis of Anthozoan GFPs to estimate the evolutionary time between each pair of species. They then used a larger dataset of six Anthozoan GFPs and species to construct an estimator that correlates sequence identity between FPs to MY of evolutionary time between the species. Based on this analysis, they estimated that esmGFP represents an equivalent of over 500 million years of evolution from the closest protein that has been found in nature. User: